An accurate identification of anatomical cardiac features allows more robust and accurate functional and structural analysis of the heart. Hence, anatomical landmark points are obviously essential for an automated cardiac MRI segmentation method. In this challenge, we focus on automatic landmark point detection method, which is particularly necessary for building a 3D LV model. These landmark points include mitral valve points, right ventricular (RV) insert points and base-to-apex central axis points
Data
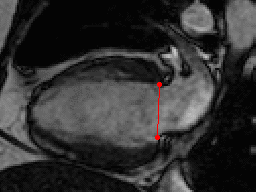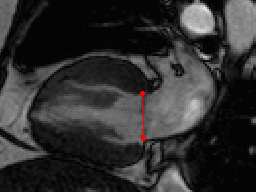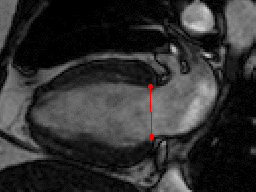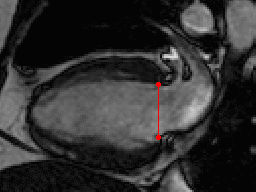
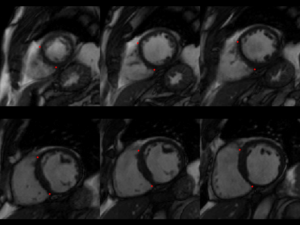
This challenge uses the same data set as in the LV Segmentation Challenge with manually annotated landmark positions were placed in the training data set as annotation data.
Note that you are not required to return your detection predictions from the test set, but if you want to validate your method, you can send us the predictions.
Data description:
| Training data size | 100 |
| Test data size | 100 |
| Annotations | 2 mitral valve points in long-axis views (all frames) |
| 1 LV center point at base in short-axis view (ED frame) | |
| 1 LV center point at apex in short-axis view (ED frame) | |
| 2 RV-LV insert points in all slices of short-axis views (all frames) | |
| Images | Steady-State Free Precession MRI in short and long axis views |
| Pathology | myocardial infarction |
Annotation Files
The cardiac landmark points are defined in a text file named as Annotations.txt. Each study has one Annotations.txt file:
PatientID: <ID>
[BASE_CENTRAL_AXIS]: 1
<X> <Y> <DICOM_filename>
[APEX_CENTRAL_AXIS]: 1
<X> <Y> <DICOM_filename>
[RV_INSERTS]: <npoints>
<X> <Y> <DICOM_filename>
<X> <Y> <DICOM_filename>
...
[MITRAL_VALVES]: <npoints>
<X> <Y> <DICOM_filename>
<X> <Y> <DICOM_filename>
...Each line contains a single landmark point with the corresponding DICOM image file. All points are in image coordinates (x, y). That means you can directly plot these points on top of the corresponding DICOM image.
For submission, please contact us by email.